3 Loading data
In this workshop we work through loading data. Once we have a curated and cleaned dataset we can work on generating insights from the data.
As a biologist you should be used to asking questions and gathering data. It is also important that you learn all aspects of the research process. This includes responsible data management (understanding data files & spreadsheet organisation, keeping data safe) and data analysis.
In this chapter we will look at the structure of data files, and how to read these with R. We will also continue to develop reproducible scripts. This means that we are writing scripts that are well organised and easy to read, and also making sure that our scripts are complete and capable of reproducing an analysis from start to finish.
Transparency and reproducibility are key values in scientific research, when you analyse data in a reproducible way it means that others can understand and check your work. It also means that the most important person can benefit from your work, YOU! When you return to an analysis after even a short break, you will be thanking your earlier self if you have worked in a clear and reproducible way, as you can pick up right where you left off.
3.1 Meet the Penguins
This data, taken from the palmerpenguins (Horst et al. (2020)) package was originally published by Gorman et al. (2014). In our course we will work with real data that has been shared by other researchers.
The palmer penguins data contains size measurements, clutch observations, and blood isotope ratios for three penguin species observed on three islands in the Palmer Archipelago, Antarctica over a study period of three years.
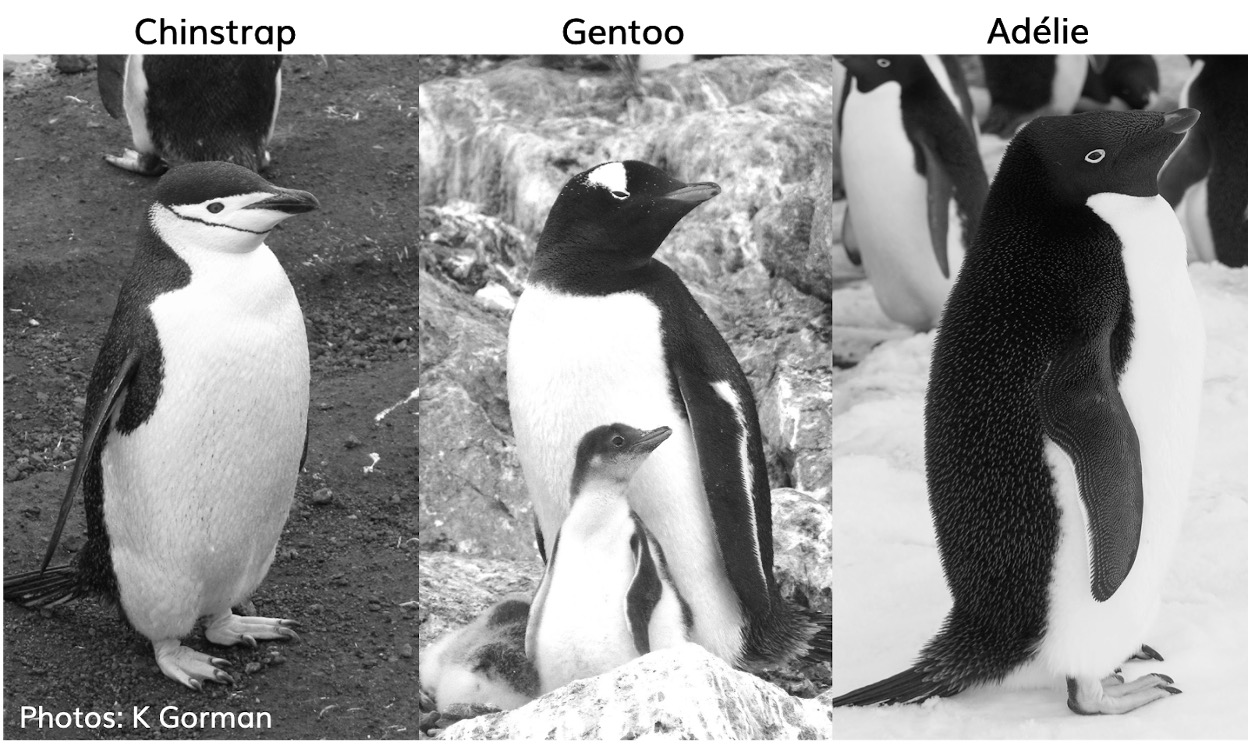
These data were collected from 2007 - 2009 by Dr. Kristen Gorman with the Palmer Station Long Term Ecological Research Program, part of the US Long Term Ecological Research Network. The data were imported directly from the Environmental Data Initiative (EDI) Data Portal, and are available for use by CC0 license (“No Rights Reserved”) in accordance with the Palmer Station Data Policy. We gratefully acknowledge Palmer Station LTER and the US LTER Network. Special thanks to Marty Downs (Director, LTER Network Office) for help regarding the data license & use. Here is our intrepid package co-author, Dr. Gorman, in action collecting some penguin data:

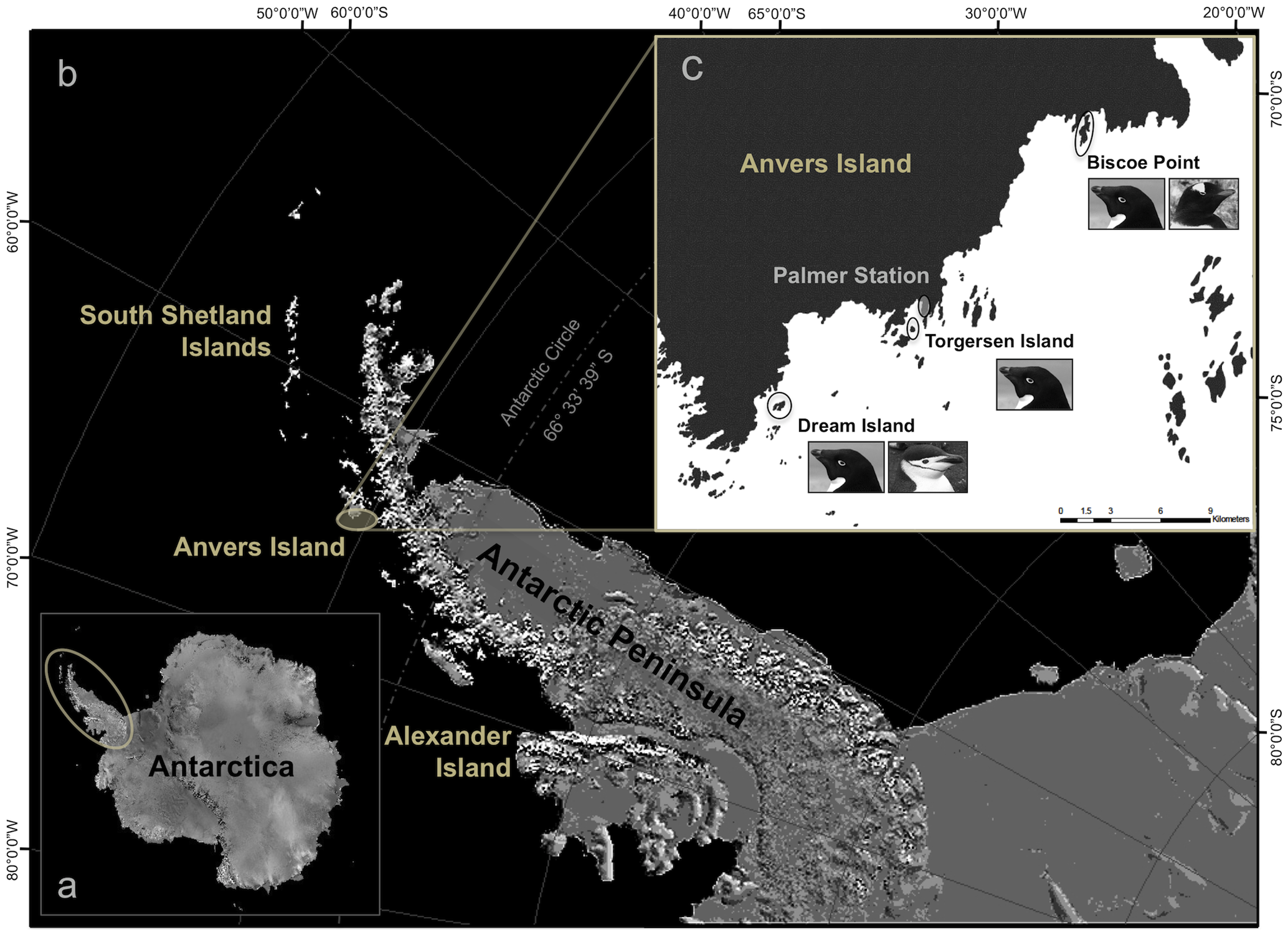
Here is a map of the study site
3.2 Activity 1: Organising our workspace
Before we can begin working with the data, we need to do some set-up.
Go to RStudio Cloud and open the
PenguinsR project-
Create the following folders using the + New Folder button in the Files tab
- data
- outputs
- scripts
R is case-sensitive so type everything exactly as printed here
Having these separate subfolders within our project helps keep things tidy, means it's harder to lose things, and lets you easily tell R exactly where to go to retrieve data.
The next step of our workflow is to have a well organised project space. RStudio Cloud does a lot of the hard work for you, each new data project can be set up with its own Project space.
We will define a project as a series of linked questions that uses one (or sometimes several) datasets. For example a coursework assignment for a particular module would be its own project, a series of linked experiments or particular research project might be its own project.
A Project will contain several files, possibly organised into sub-folders containing data, R scripts and final outputs. You might want to keep any information (wider reading) you have gathered that is relevant to your project.
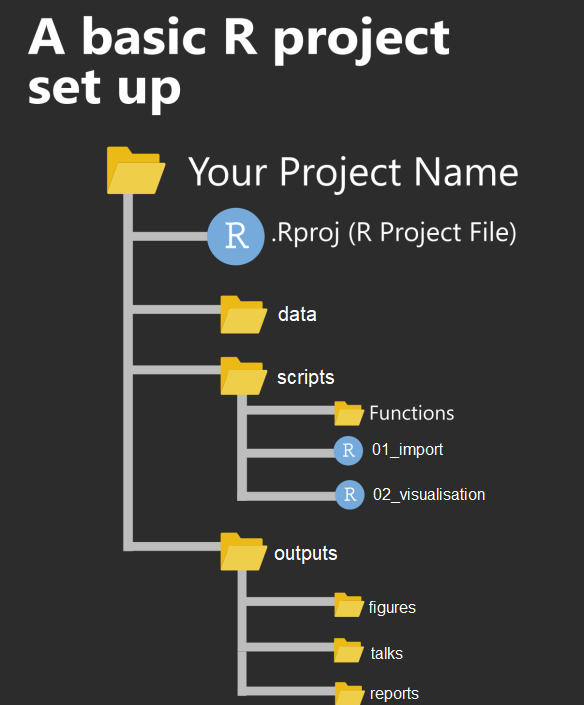
Figure 3.1: An example of a typical R project set-up
Within this project you will notice there is already one file .Rproj. This is an R project file, this is a very useful feature, it interacts with R to tell it you are working in a very specific place on the computer (in this case the cloud server we have dialed into). It means R will automatically treat the location of your project file as the 'working directory' and makes importing and exporting easier1.
3.3 Activity 2: Access our data
Now that we have a project workspace, we are ready to import some data.
Use the link below to open a page in your browser with the data open
Right-click Save As to download in csv format to your computer (Make a note of where the file is being downloaded to e.g. Downloads)
Compare how the data looks in "raw" format to when you open the same data with Excel
At first glance the data might look quite strange and messy. It has been stored as a CSV or comma-separated values file. CSV files are plain text files that can store large amounts of data, and can readily be imported into a spreadsheet or storage database.
These files are the simplest form of database, no coloured cells, no formulae, no text formatting. Each row is a row of the data, each value of a row (previously separate columns) is separated by a comma.
This file format helps us maintain an ethos Keep Raw Data Raw -
In many cases, the captured or collected data may be unique and impossible to reproduce, such as measurements in a lab or field observations. For this reason, they should be protected from any possible loss. Every time a change is made to a raw data file it threatens the integrity of that information.
In practice, that means we only use our data file for data entry and storage. All the data manipulation, cleaning and analysis happens in R, using transparent and reproducible scripts.
We avoid saving files in the Excel format because they have a nasty habit of formatting or even losing data when the file gets large enough.
If you need to add data to a csv file, you can always open it in an Excel-like program and add more information, but remember to save it in the original csv format afterwards.

Figure 3.2: Top image: Penguins data viewed in Excel, Bottom image: Penguins data in native csv format
In raw format, each line of a CSV is separated by commas for different values. When you open this in a spreadsheet program like Excel it automatically converts those comma-separated values into tables and columns.
You are probably more used to working with Excel (.xls and .xlsx) file formats, but while these are widely supported, CSV files, as simple text formats are supported by ALL data interfaces. They are also not proprietary (e.g. the Excel format is owned by Microsoft), so by working with a .csv format your data is more open and accessible.
3.4 Activity 3: Upload our data
The data is now in your Downloads folder on your computer
We need to upload the data to our remote cloud-server (RStudio Cloud), select the upload files to server button in the Files tab
Put your file into the data folder - if you make a mistake select the tickbox for your file, go to the cogs button and choose the option Move.
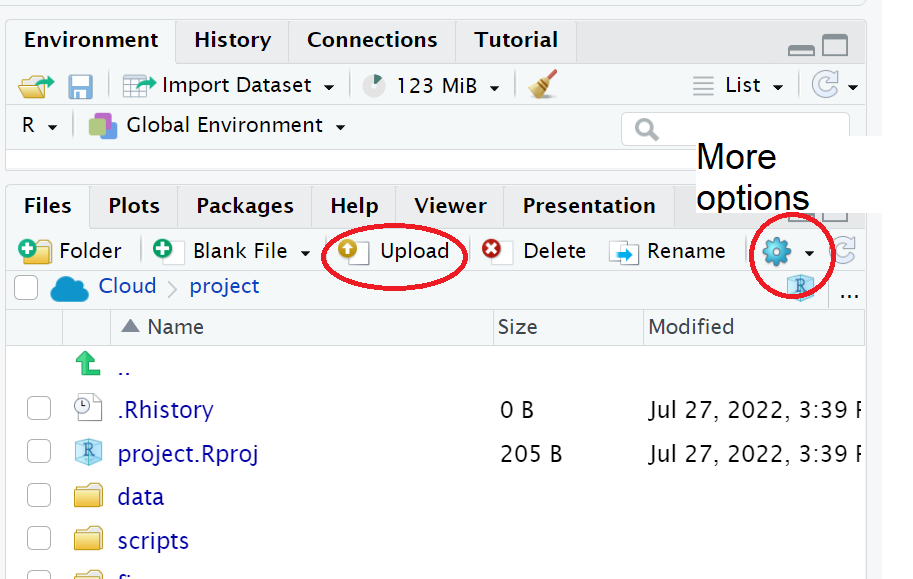
Figure 3.3: Highlighted the buttons to upload files, and more options
3.5 Activity 4: Make a script
Let's now create a new R script file in which we will write instructions and store comments for manipulating data, developing tables and figures. Use the File > New Script menu item and select an R Script.
Add the following:
#___________________________----
# SET UP ----
## An analysis of the bill dimensions of male and female Adelie, Gentoo and Chinstrap penguins ----
### Data first published in Gorman, KB, TD Williams, and WR Fraser. 2014. “Ecological Sexual Dimorphism and Environmental Variability Within a Community of Antarctic Penguins (Genus Pygoscelis).” PLos One 9 (3): e90081. https://doi.org/10.1371/journal.pone.0090081. ------
#__________________________----Then load the following add-on package to the R script, just underneath these comments. Tidyverse isn't actually one package, but a bundle of many different packages that play well together - for example it includes ggplot2 which we used in the last session, so we don't have to call that separately
Add the following to your script:
# PACKAGES ----
library(tidyverse) # tidy data packages
library(janitor) # cleans variable names
library(lubridate) # make sure dates are processed properly
#__________________________----Save this file inside the scripts folder and call it 01_import_penguins_data.R
Click on the document outline button (top right of script pane). This will show you how the use of
#TITLES----
Allows us to build a series of headers and subheaders, this is very useful when using longer scripts.
3.6 Activity 5: Read in data
Now we can read in the data. To do this we will use the function read_csv() that allows us to read in .csv files. There are also functions that allow you to read in .xlsx files and other formats, however in this course we will only use .csv files.
First, we will create an object called
penguins_datathat contains the data in thepenguins_raw.csvfile.Add the following to your script, and check the document outline:
# IMPORT DATA ----
penguins <- read_csv ("data/penguins_raw.csv")
head(penguins) # check the data has loaded, prints first 10 rows of dataframe
#__________________________----
There is also a function called read.csv(). Be very careful NOT to use this function instead of read_csv() as they have different ways of naming columns.
3.7 Filepaths
In the example above the read_csv() function requires you to provide a filepath (in "quotes"), in order to tell R where the file you wish to read is located in this example there are two components
"data/" - specifies the directory in which to look for the file
"penguins_raw.csv" - specifies the name and format of the file
3.7.1 Directories
A directory refers to a folder on a computer that has relationships to other folders. The term “directory” considers the relationship between that folder and the folders within and around it. Directories are hierarchical which means that they can exist within other folders as well as have folders exist within them.
No idea what directories or files are? You are not alone File not Found
A "parent" directory is any folder that contains a subdirectory. For example your downloads folder is a directory, it is the parent directory to any subdirectories or files contained within it.
3.7.2 Home directory
The home directory on a computer is a directory defined by your operating system. The home directory is the primary directory for your user account on your computer. Your files are by default stored in your home directory.
On Windows, the home directory is typically
C:\Users\your-username.On Mac and Linux, the home directory is typically
/home/your-username.
3.7.3 Working directory
The working directory refers to the directory on your computer that a tool assumes is the starting place for all filepaths
3.7.4 Absolute vs Relative filepaths
What has this got to do with working in R?
When you use any programming language, you have to specify filepaths in order for the program to find files to read-in or where to output files.
An Absolute file path is a path that contains the entire path to a file or directory starting from your Home directory and ending at the file or directory you wish to access e.g.
/home/your-username/project/data/penguins_raw.csv
The main drawbacks of using absolute file paths are:
If you share files, another user won’t have the same directory structure as you, so they will need to recreate the file paths
if you alter your directory structure, you’ll need to rewrite the paths
an absolute file path will likely be longer than a relative path, more of the backslashes will need to be edited, so there is more scope for error.
As different computers can have different path constructions, any scripts that use absolute filepaths are not very reproducible.
A Relative filepath is the path that is relative to the working directory location on your computer.
When you use RStudio Projects, wherever the .Rproj file is located is set to the working directory. This means that if the .Rproj file is located in your project folder then the relative path to your data is:
data/penguins_raw.csv
This filepath is shorter and it means you could share your project with someone else and the script would run without any editing.
For those of you using RStudio Cloud, remember you are working on a Linux OS cloud server, each of you will have a different absolute filepath - but the scripts for the project you are working on right now work because you are using relative filepaths
3.8 Activity 5: Check your script
#___________________________----
# SET UP ----
## An analysis of the bill dimensions of male and female Adelie, Gentoo and Chinstrap penguins ----
### Data first published in Gorman, KB, TD Williams, and WR Fraser. 2014. “Ecological Sexual Dimorphism and Environmental Variability Within a Community of Antarctic Penguins (Genus Pygoscelis).” PLos One 9 (3): e90081. https://doi.org/10.1371/journal.pone.0090081. ------
#__________________________----
# PACKAGES ----
library(tidyverse) # tidy data packages
library(janitor) # cleans variable names
library(lubridate) # make sure dates are processed properly
#__________________________----
# IMPORT DATA ----
penguins <- read_csv ("data/penguins_raw.csv")
head(penguins) # check the data has loaded, prints first 10 rows of dataframe
#__________________________----3.9 Activity 7: Test yourself
Question 1. In order to make your R project reproducible what filepath should you use?
Question 2. Which of these would be acceptable to include in a raw datafile?
Question 3. What should always be the first set of functions in our script? ?()
Question 4. When reading in data to R we should use
Question 5. What format is the penguins data in?
Each column is a unique variable and each row is a unique observation so this data is in a long (tidy) format
Question 6. The working directory for your projects is by default set to the location of?
Question 7. Using the filepath "data/penguins_raw.csv" is an example of
Question 8. What operator do I need to use if I wish to assign the output of the read_csv function to an R object (rather than just print the dataframe into the console)?